Kepler Achieves State-of-the-Art Performance on BixBench Computational Biology Benchmark
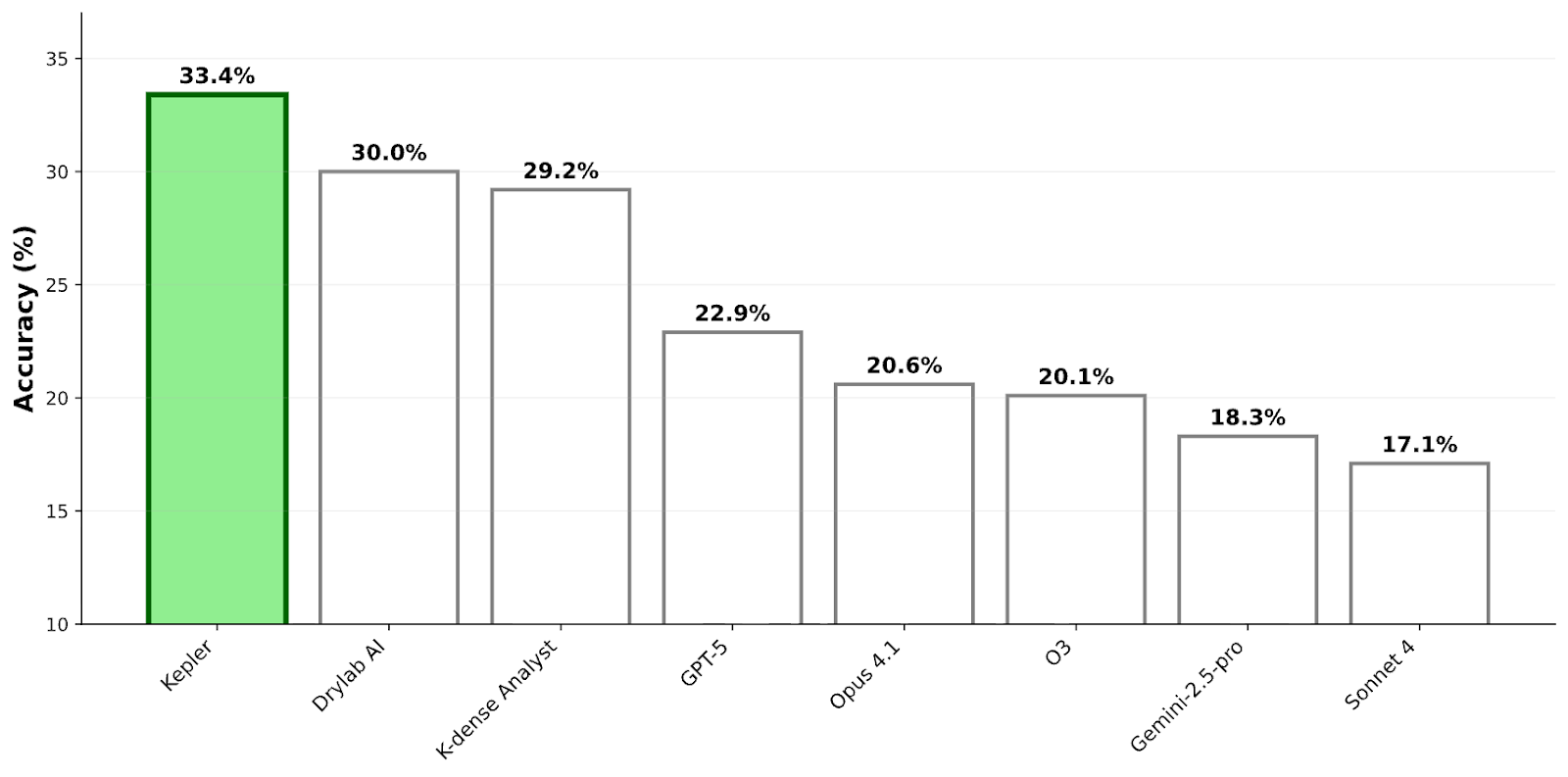
Today we are excited to announce Kepler has achieved a state-of-the-art 33.4% accuracy on BixBench — representing the highest score among all general and domain-specific agents evaluated to date.
In this post, we describe:
- The Bixbench benchmark design
- How we ran Kepler on Bixbench and the results
- Discussion around benchmark design and evaluation
- Some real-world examples of how bioinformatics is done interactively with Kepler
What is BixBench?
BixBench is a comprehensive benchmark developed by FutureHouse and ScienceMachine to evaluate the capabilities of AI agents in real-world bioinformatics tasks. Unlike traditional benchmarks that focus on facts recall or simple coding tasks, BixBench presents agents with authentic biological data analysis scenarios that mirror the complex, multi-step analytical workflows that bioinformaticians encounter daily.
The benchmark comprises 53 real-world analytical scenarios paired with 296 open-answer questions, mostly mined from existing papers, that require agents to:
- Explore heterogeneous biological datasets
- Navigate complex data structures and choose appropriate analytical approaches
- Perform sophisticated multi-step computational analyses
- Interpret nuanced results in the context of biological hypotheses
Each "capsule" in BixBench contains actual research data, a guiding hypothesis, and 3-7 associated questions that can only be answered through rigorous data exploration and analysis (not by recall). The benchmark covers diverse areas of computational biology including genomics, transcriptomics, phylogenetics, imaging, and functional genomics — making it one of the most comprehensive evaluations of AI capabilities in biological data science.
In fact, even successfully running BixBench is quite challenging. The authors used the Aviary agent framework and a custom Docker environment developed at FutureHouse. The necessary components include:
- Terminal use - the ability to navigate complex directory structures of source data
- Bioinformatics compute environment in both Python and R - domain-specific packages for genomics, proteomics, and statistical analysis, e.g. Bioconductor, scikit-learn
- Jupyter notebook execution (or equivalent) - the capability to iteratively develop, execute, and debug code cells while maintaining state across analysis steps
- Data format handling - proficiency with diverse data formats including CSV, XLSX, RDS, FASTA, TIFF, VCF, etc
- Bioinformatics domain knowledge - genome variant analysis, gene ontology and pathway analysis, differential expression workflows, phylogenetic analysis, etc.
- Statistical reasoning - understanding of appropriate statistical tests, multiple testing correction, and interpretation of p-values and effect sizes in biological contexts
- Visualization and multimodal reasoning - generating publication-quality plots and figures, and the ability to interpret those plots to determine next steps
Kepler’s results on BixBench
Kepler addresses these challenges through two integrated components: the Kepler Agent Runtime — a highly scalable, bioinformatics-curated environment with specialized tools — and the Kepler Agent itself, designed to closely interact with this runtime to conduct experiments according to user intent.
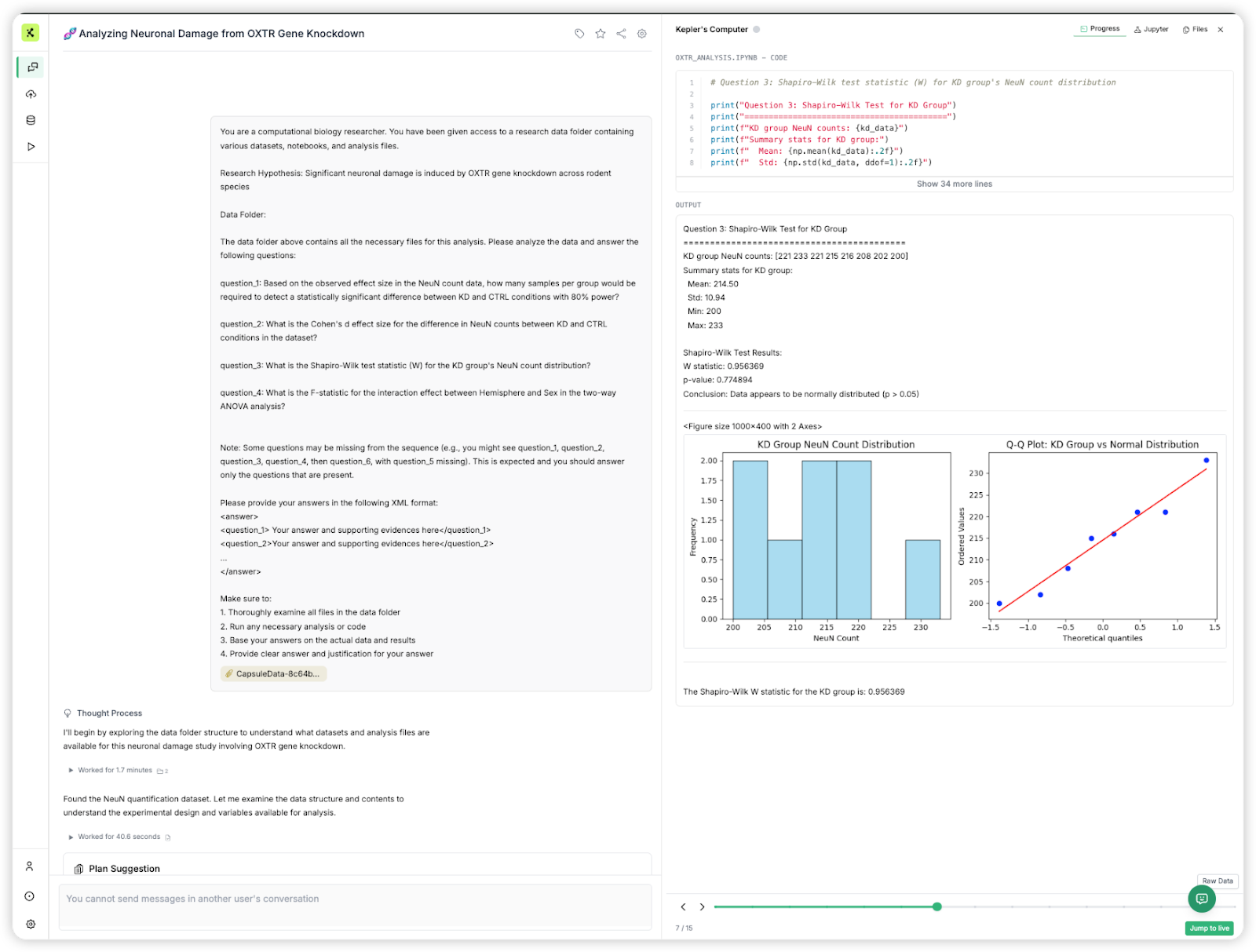
What sets Kepler apart is not just its performance, but its approach to analysis. The agent provides systematic analysis planning, detailed step-by-step execution, and transparent thought processes that mirror how expert bioinformaticians approach complex data challenges. Importantly, we evaluated Kepler on the exact same platform that our users access, ensuring that benchmark performance translates directly to real-world user experience. Notably, whereas approaches such as K-Dense Analyst used a dual-loop (planning, orchestrator) design with ten specialized agents, for the purposes of Bixbench evaluation we only used a simple single-agent architecture (see more about this topic on Cognition’s blog here).
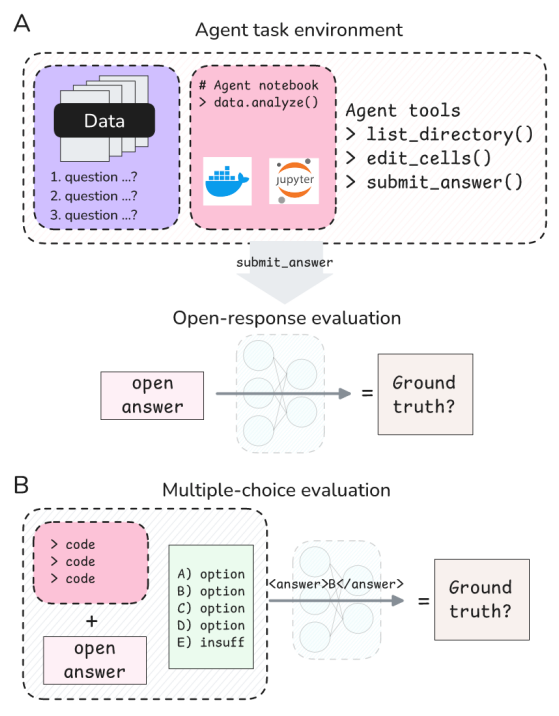
Our evaluation follows the BixBench methodology under the Open Answer paradigm, similar to K-Dense Analyst (the methodology for the Multiple Choice Question paradigm was unclear to us, and therefore it was not implemented):
- Kepler receives only the input data, research hypothesis and 3-7 question texts— no access to ground truth or the multiple choice options (See Figure 2 with the exact prompt that Kepler receives).
- Kepler submits the final answer to each question along with an explanation at the end of the analysis
- A separate LLM-as-a-judge system (prompt linked in the appendix) using GPT-5 is given the questions texts, ground truth answers, Kepler’s answers to each question along with the explanation, and categorizes each answer as correct (1) or incorrect (0), allowing a small epsilon for rounding errors.


Full list of results and scores for each capsule can be found here.
Issues with Benchmark Design and Evaluation
We want to preface this by saying that it is by definition difficult to evaluate open-ended data science analyses, and applaud the authors efforts in coming up with a well thought out paradigm with quantifiable results.
That being said, there are many parts of this benchmark design that could be improved:
Lack of Human Baseline: As the authors have stated, it is important to have another set of humans perform the same tasks with access to the same information that the agents have access to to establish a baseline of performance. The original authors stated that “As the seed capsules used to generate BixBench tasks were generated de novo by such human experts, we anticipate that additional human experts would perform significantly higher than the agent performance reported here, and thus did not prioritize gathering this data.” It is actually not evident that a second set of humans would perform well, since different bioinformaticians routinely interpret the same questions in different ways.
For example, in the question bix-10, where Kepler got 0 questions correct, the research hypothesis was "Tubercolosis BCG re-vaccination reduces COVID-19 disease severity among healthcare workers". However, upon detailed analysis, Kepler's approach was actually likely more correct than the human benchmark. The human analyst seems to have misunderstood the research question, analyzing all adverse events across 791 patients rather than focusing specifically on COVID-19 cases. This led them to conclude that BCG vaccination was harmful (OR=1.524), when in reality they were measuring general adverse events including expected vaccination side effects. In contrast, Kepler correctly isolated the 174 COVID-19 cases from the adverse events data and found that BCG showed protective trends against COVID-19 severity (OR=0.637).
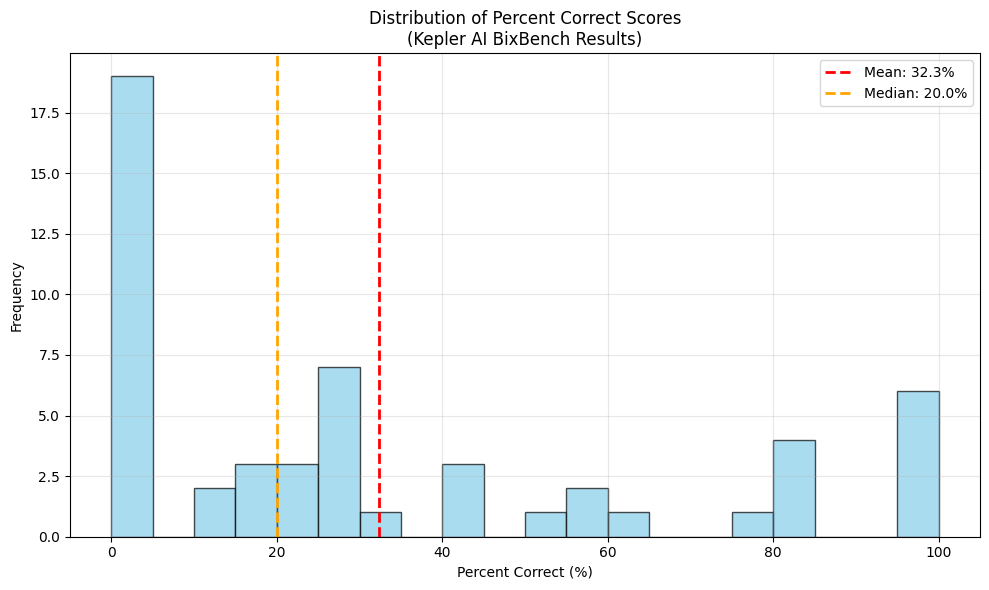
Looking at the distribution of failures on a capsule level (Figure 6), 19 tasks (35.8%) had 0 questions correct, suggesting a fundamental misunderstanding or misspecification of the analysis questions or goals. A human baseline here would greatly help with sorting these errors into different categories.
Backward Engineering: Tasks were created by having humans first complete open-ended analyses in a notebook, and then Claude 3.5 Sonnet was used to generate questions from the notebooks post-hoc. Since the original analysis was open-ended and presumably many analysis trajectories were possible, forcing a series of related questions on an unknown trajectory may not be reasonable.
Real-world interactive bioinformatics with Kepler
For the purposes of running Bixbench programmatically, we disabled many features in Kepler that allow interaction with the user, however we believe that this interaction is critical. In fact, most users do not know how to prompt AI to come up with concrete analysis plans, and users will need to learn prompt engineering just like how software engineers learned how to use coding agents. Kepler is designed to work with the user to iterate back and forth to allow for a more concrete plan. During the analyses, Kepler also flags issues with the data, and confers with the users about forks in the road during an analysis, where many different paths may be valid, depending on the user’s goals.
In this example, the user uses Kepler to search for GEO studies with bulk RNA-seq data on HNF1A knockout cells, Kepler helps selects an appropriate dataset, does QC, flags potential issues, and asks the user what analysis to do at each step given the current information.
In this example, the user starts out with an exploratory question about the possibilities of analyses within DepMap, and then narrows down to a specific question.
Bioinformatics in the real world also involves frequent references to external datasets and literature, so the data that is necessary for an analysis is not necessarily available at the start. Kepler automatically discovers useful tools and resources, and checks any results with literature search.
Conclusion
This is just the beginning! We are energized by the emerging interest in AI agents in science, and will be integrating more benchmarks, as well as producing our own benchmarks soon to contribute to the community. Even though what is now achievable with AI would be unthinkable just a year ago, there is still a long way to go before AI agents can do end-to-end scientific discovery, and we are excited to contribute to this mission.
Appendix:
User prompt template:

LLM judge prompt:


Connect the dots with Kepler
Try out Kepler today, or book a call with us about your organization's use case
Connect the dots with Kepler
Try out Kepler today, or book a call with us about your organization's use case

